import copy, math
import numpy as np
%matplotlib widget
import matplotlib.pyplot as plt
from lab_utils_common import dlc, plot_data, plt_tumor_data, sigmoid, compute_cost_logistic
from plt_quad_logistic import plt_quad_logistic, plt_prob
plt.style.use('./deeplearning.mplstyle')Gradient Descent
So how do we find the values for w and b to minimize the cost for the model?
Recall from previous work that we have to iterate over the entire training set and repeat while updating w & b simulteanously
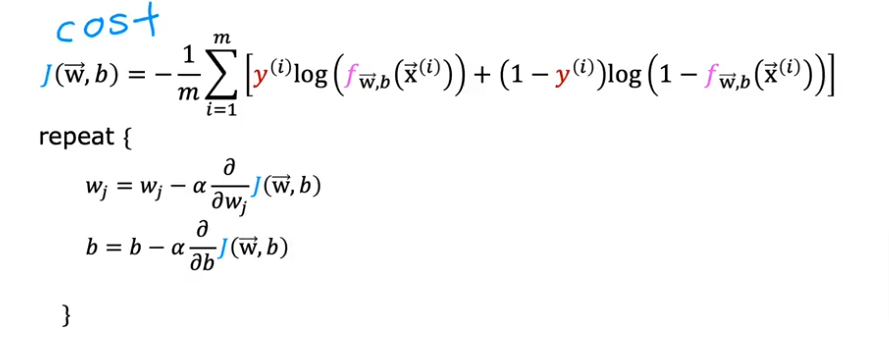
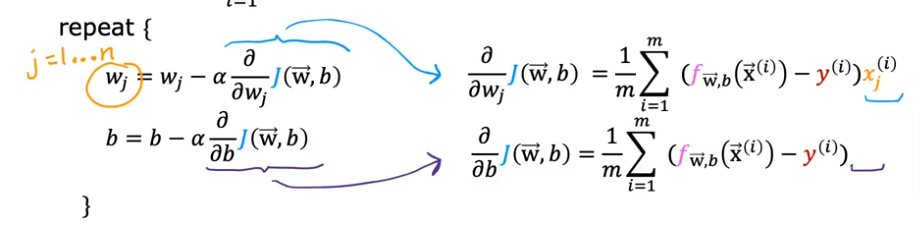
summarized as the same formula we used for linear regression
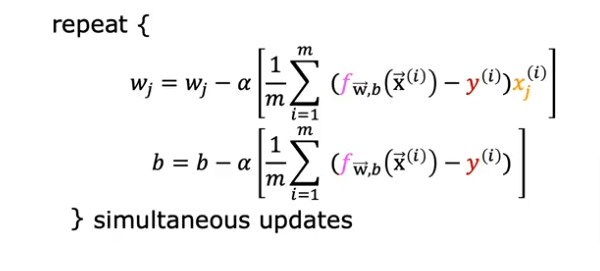
Yes except that for a logistic regression
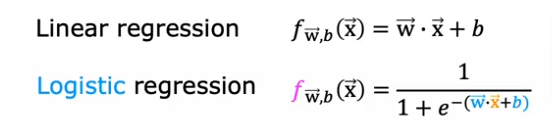
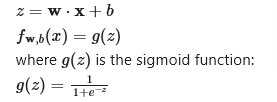
Code
X_train = np.array([[0.5, 1.5], [1,1], [1.5, 0.5], [3, 0.5], [2, 2], [1, 2.5]])
y_train = np.array([0, 0, 0, 1, 1, 1])fig,ax = plt.subplots(1,1,figsize=(4,4))
plot_data(X_train, y_train, ax)
ax.axis([0, 4, 0, 3.5])
ax.set_ylabel('$x_1$', fontsize=12)
ax.set_xlabel('$x_0$', fontsize=12)
plt.show()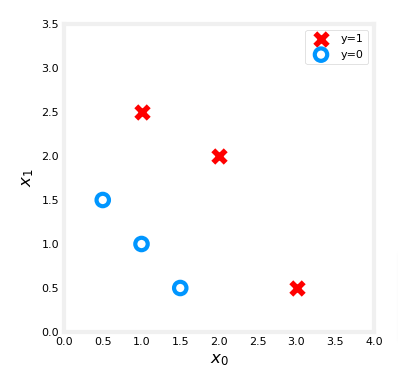
Gradient Descent
def compute_gradient_logistic(X, y, w, b):
"""
Computes the gradient for logistic regression
Args:
X (ndarray (m,n): Data, m examples with n features
y (ndarray (m,)): target values
w (ndarray (n,)): model parameters
b (scalar) : model parameter
Returns
dj_dw (ndarray (n,)): The gradient of the cost w.r.t. the parameters w.
dj_db (scalar) : The gradient of the cost w.r.t. the parameter b.
"""
m,n = X.shape
dj_dw = np.zeros((n,)) #(n,)
dj_db = 0.
for i in range(m):
f_wb_i = sigmoid(np.dot(X[i],w) + b) #(n,)(n,)=scalar
err_i = f_wb_i - y[i] #scalar
for j in range(n):
dj_dw[j] = dj_dw[j] + err_i * X[i,j] #scalar
dj_db = dj_db + err_i
dj_dw = dj_dw/m #(n,)
dj_db = dj_db/m #scalar
return dj_db, dj_dw X_tmp = np.array([[0.5, 1.5], [1,1], [1.5, 0.5], [3, 0.5], [2, 2], [1, 2.5]])
y_tmp = np.array([0, 0, 0, 1, 1, 1])
w_tmp = np.array([2.,3.])
b_tmp = 1.
dj_db_tmp, dj_dw_tmp = compute_gradient_logistic(X_tmp, y_tmp, w_tmp, b_tmp)
print(f"dj_db: {dj_db_tmp}" )
print(f"dj_dw: {dj_dw_tmp.tolist()}" )Gradient Descent
def gradient_descent(X, y, w_in, b_in, alpha, num_iters):
"""
Performs batch gradient descent
Args:
X (ndarray (m,n) : Data, m examples with n features
y (ndarray (m,)) : target values
w_in (ndarray (n,)): Initial values of model parameters
b_in (scalar) : Initial values of model parameter
alpha (float) : Learning rate
num_iters (scalar) : number of iterations to run gradient descent
Returns:
w (ndarray (n,)) : Updated values of parameters
b (scalar) : Updated value of parameter
"""
# An array to store cost J and w's at each iteration primarily for graphing later
J_history = []
w = copy.deepcopy(w_in) #avoid modifying global w within function
b = b_in
for i in range(num_iters):
# Calculate the gradient and update the parameters
dj_db, dj_dw = compute_gradient_logistic(X, y, w, b)
# Update Parameters using w, b, alpha and gradient
w = w - alpha * dj_dw
b = b - alpha * dj_db
# Save cost J at each iteration
if i<100000: # prevent resource exhaustion
J_history.append( compute_cost_logistic(X, y, w, b) )
# Print cost every at intervals 10 times or as many iterations if < 10
if i% math.ceil(num_iters / 10) == 0:
print(f"Iteration {i:4d}: Cost {J_history[-1]} ")
return w, b, J_history #return final w,b and J history for graphingCalculate GD
w_tmp = np.zeros_like(X_train[0])
b_tmp = 0.
alph = 0.1
iters = 10000
w_out, b_out, _ = gradient_descent(X_train, y_train, w_tmp, b_tmp, alph, iters)
print(f"\nupdated parameters: w:{w_out}, b:{b_out}")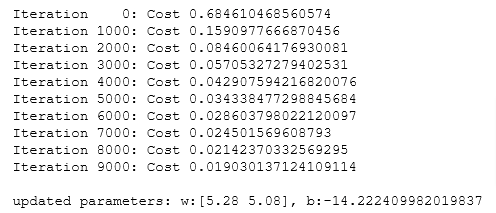
Plot
fig,ax = plt.subplots(1,1,figsize=(5,4))
# plot the probability
plt_prob(ax, w_out, b_out)
# Plot the original data
ax.set_ylabel(r'$x_1$')
ax.set_xlabel(r'$x_0$')
ax.axis([0, 4, 0, 3.5])
plot_data(X_train,y_train,ax)
# Plot the decision boundary
x0 = -b_out/w_out[0]
x1 = -b_out/w_out[1]
ax.plot([0,x0],[x1,0], c=dlc["dlblue"], lw=1)
plt.show()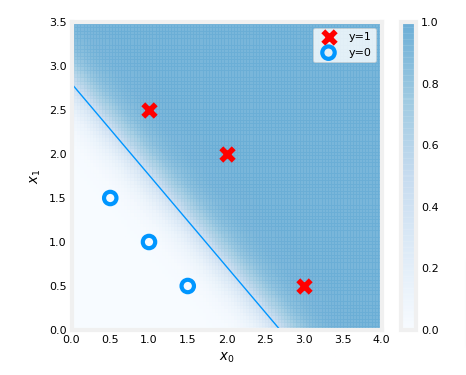
In the plot above:
- the shading reflects the probability y=1 (result prior to decision boundary)
- the decision boundary is the line at which the probability = 0.5
One Variable Dataset
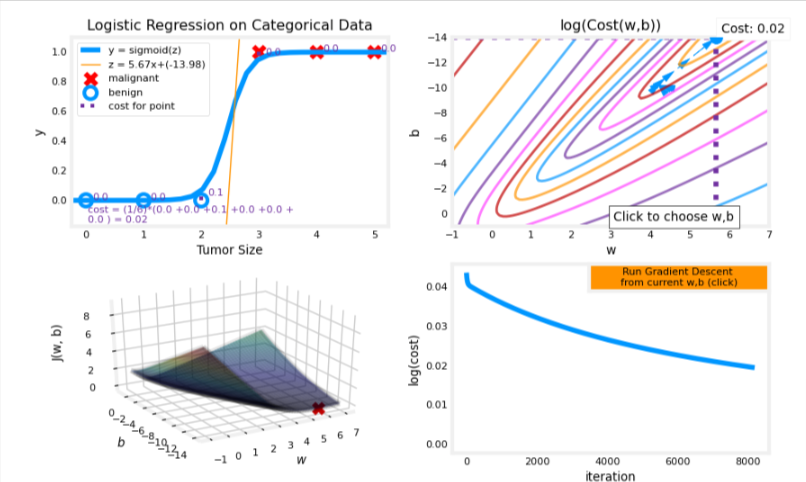
Let’s return to a one-variable data set. With just two parameters, 𝑤w, 𝑏b, it is possible to plot the cost function using a contour plot to get a better idea of what gradient descent is up to.
x_train = np.array([0., 1, 2, 3, 4, 5])
y_train = np.array([0, 0, 0, 1, 1, 1])The data points with label 𝑦=1y=1 are shown as red crosses, while the data points with label 𝑦=0y=0 are shown as blue circles.
fig,ax = plt.subplots(1,1,figsize=(4,3))
plt_tumor_data(x_train, y_train, ax)
plt.show()
In the plot below, try:
- changing 𝑤w and 𝑏b by clicking within the contour plot on the upper right.
- changes may take a second or two
- note the changing value of cost on the upper left plot.
- note the cost is accumulated by a loss on each example (vertical dotted lines)
- run gradient descent by clicking the orange button.
note the steadily decreasing cost (contour and cost plot are in log(cost)
clicking in the contour plot will reset the model for a new run
- to reset the plot, rerun the cell
w_range = np.array([-1, 7])
b_range = np.array([1, -14])
quad = plt_quad_logistic( x_train, y_train, w_range, b_range )